Immunology &
Autoimmune Disorders
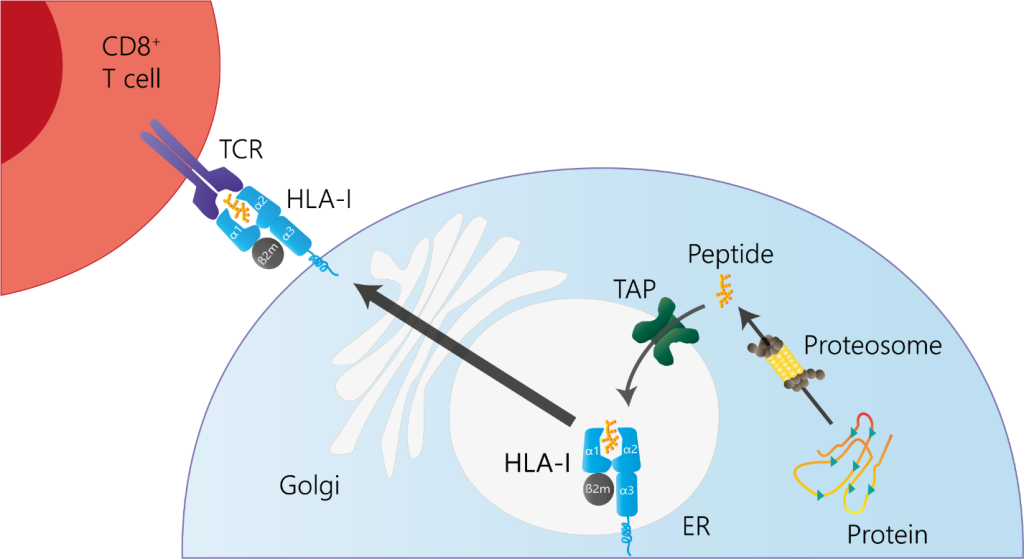
Figure 1. Schematic overview of HLA-I antigen presentation to CD8⁺ cells.
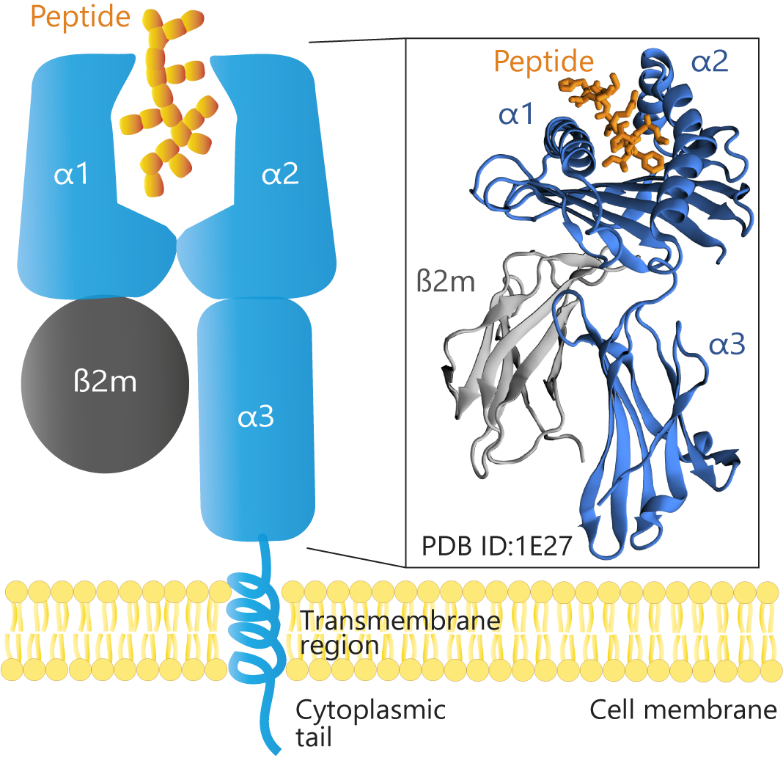
Figure 2. Schematic and structural representation of HLA-B*51 (PDB ID: 1E27) loaded with a peptide. The α1 and α2 domains form the peptide-binding groove, and the α3 domain lies closer to the cell membrane, anchoring the complex in conjunction with the transmembrane region. β2-microglobulin (β2m) provides structural support by stabilizing the heavy chain. The cytoplasmic tail extends into the cell’s interior. This organization underlies HLA-B*51’s critical role in presenting antigens and mediating immune responses.
Video 1. In silico pulling experiment. Soft-spring steered MD simulation mimicking AFM conditions to measure rupture forces between the peptide and HLA-B*51.
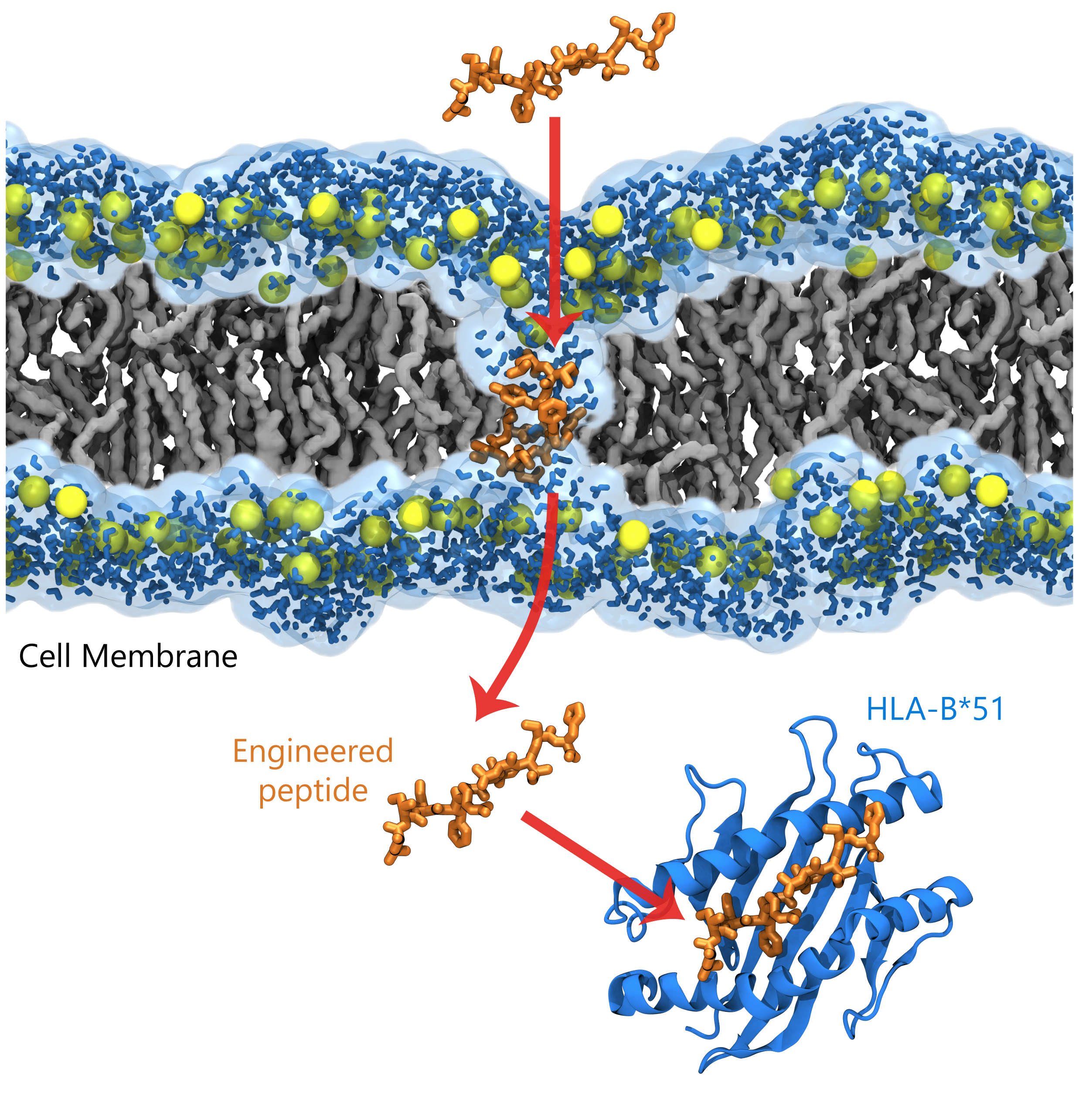
Figure 3. A Gur lab-designed peptide engineered to translocate into the cell membrane and exhibit high binding affinity for HLA-B*51. The top-left panel illustrates the peptide translocating through the lipid bilayer, whereas the bottom-left panel shows the peptide in its bound conformation with HLA-B*51.
Our Published Findings
Gur, M., Golcuk, M., Gul, A., Erman, B. (2020). Molecular dynamics simulations provide molecular insights into the role of HLA-B51 in Behçet’s disease pathogenesis. Chemical Biology & Drug Design, 96(1), 644-658 (5-year IF: 2.9, IF: 3.2)
Past Funding
Tubitak (1001, Grant No: 119Z553)
Lab Members Involved
Sema Zeynep Yilmaz, Mert Golcuk, Derman Basturk
Developing Next-Generation Peptide-Based Therapeutics for Autoimmune Diseases
Major histocompatibility complex (MHC) proteins, known as human leukocyte antigens (HLA) in humans, present protein fragments (peptides) on the cell surface. These fragments originate from inside the cell for HLA class I and from outside sources for HLA class II. HLA class I molecules, such as HLA-A, HLA-B, and HLA-C, bind peptides that come from proteins degraded within the cell, often from viruses or cancerous changes (Figure 1 and 2). After being transported into the endoplasmic reticulum (ER), these peptides are loaded onto HLA-I molecules and then displayed on the cell surface, allowing CD8+ T cells to inspect them. The T cells use their receptors to recognize the shape formed by both the peptide and the HLA molecule. Subtle differences in the peptide sequence can alter this interaction, determining whether the T cell initiates an immune response.
Because HLA variants influence how the immune system distinguishes between self and non-self, certain HLA alleles are strongly associated with specific diseases. One notable example is the HLA-B*51 allele, which is commonly found in patients with Behçet’s disease (a condition characterized by recurring inflammation of the mouth, genitals, skin, and eyes). The frequent presence of HLA-B*51 in individuals with Behçet’s disease suggests that specific HLA variants can play a significant role in driving abnormal immune responses, contributing to disease development.
HLA variants also influence the susceptibility and course of other inflammatory conditions, such as Takayasu’s arteritis. This disease predominantly affects the aorta and its major branches, leading to inflammation (vasculitis) and narrowing (stenosis) of these large vessels. The HLA-B*52 allele has been linked to an increased risk of Takayasu’s arteritis, suggesting that certain HLA subtypes can predispose individuals to specific immune-mediated vascular diseases. Much like the association seen in Behçet’s disease, the presence of HLA-B*52 may alter the immune system’s response to self-peptides, ultimately contributing to the excessive inflammation and damage characteristics of Takayasu’s arteritis.
Yet, the precise molecular mechanisms linking these HLA variants to disease remain only partially understood, and effective therapeutics specifically targeting HLA molecules have yet to be developed for these conditions. Consequently, no definitive cure exists for Behçet’s disease, Takayasu’s arteritis, or related disorders. To address these challenges, my laboratory is taking a two-pronged approach: i) investigating the fundamental molecular events that underpin HLA-associated pathogenicity, and ii) exploring the design of novel peptide-based therapeutics aimed at modulating these molecular interactions. Through this work, our goal is to prevent the formation of pathogenic HLA–peptide complexes and thereby mitigate the resulting aberrant immune responses.
In pursuit of these goals, we are methodically dissecting the interaction and binding dynamics between HLA-I molecules and both self-derived and viral peptides, as well as their subsequent recognition by the T-cell receptor (TCR). By mapping these molecular interactions in detail, we aim to identify the critical amino acid residues and allosteric pathways that govern binding affinity, specificity, and the precise regulation of these immune interfaces. Ultimately, this knowledge has the potential to guide the development of innovative strategies to modulate HLA–peptide–TCR interactions and mitigate pathogenic immune responses.
We have uncovered several previously uncharacterized molecular mechanisms of HLA-B*51, including its looser peptide-binding dynamics compared to HLA-B*52, shedding light on its involvement in Behçet’s disease pathogenesis. Building on these findings, we are leading a research effort focused on determining the molecular features that govern HLA-B*51’s interactions with its self-peptides (endogenous peptides derived from the body’s own proteins). To achieve this, we employ exhaustive, unbiased molecular dynamics (MD) simulations and then apply statistical mechanics and machine learning techniques to analyze the resulting data. This approach provides unique insights that are not readily attainable through conventional methods.
In addition, we perform in silico “pulling” experiments using steered MD (SMD) simulations designed to mimic the conditions of high-speed atomic force microscopy (AFM). These simulations utilize loading rates one to two orders of magnitude lower than standard SMD protocols, rendering them not only more computationally demanding but also more realistic. During SMD simulations, a virtual “dummy atom” is linked to the molecule of interest via a virtual spring. We conduct two types of SMD simulations, both employing constant-velocity pulling. In the first type, a stiff spring ensures that the pulled atoms closely follow the dummy atom, satisfying the “stiff spring approximation” and enabling us to generate free energy profiles (potentials of mean force) for the unbinding process. In the second type, a softer spring replicates the mechanical properties of the AFM lever, allowing us to estimate rupture forces (Video 1) accurately. By measuring both rupture forces and binding affinities, we obtain data directly comparable to experimental results along with molecular details about the binding/unbinding process not visible to experimental techniques, further enhancing our understanding of HLA-peptide interactions and their implications in disease.
Building on our atomic-level insights into HLA–peptide interactions and their role in disease pathology, our laboratory is focusing on engineering novel therapeutic peptides. We harness the biophysical properties of naturally occurring peptide scaffolds, combine them with our in-house design methods, and apply simulation and AI/ML-driven de novo peptide engineering to achieve enhanced HLA-I binding and optimized cell membrane translocation capabilities. By systematically evaluating the binding affinity of these peptides to HLA-B*51 and assessing their membrane translocation in silico, we can benchmark them against naturally occurring peptides. This approach lays the groundwork for new classes of peptide therapeutics designed to more precisely target autoimmune diseases. In contrast to broad immunosuppressive therapies, such engineered peptide treatments—particularly those targeting HLA-B*51—could offer more specific interventions for Behçet’s disease and Takayasu’s arteritis, reducing side effects and potentially advancing the treatment landscape for these and related immune-mediated conditions.